]. "Proletarian of mental labor." Physicist by education. I work in the field of processing medical and biological information 30+ years.
I’ve been working in R for exactly 10 years, having migrated to it after 15 years of close collaboration with Matlab. The primary reason for migrating to another work platform was my own physical migration to the opposite end of the Earth in Auckland, New Zealand. Here life from the first days pushed me into the arms of R, which I have not yet had to regret.
More and more often I see outbreaks of interest in R in the professional network. Well, articles about him appear on this respected resource. Further below the cut is my first attempt at a Russian-language introduction to R - the first (verbal) part of the presentation that I did for colleagues from the Animal Science Faculty, Iova State University three years ago.
(to the side: but it turns out that it’s difficult to translate yourself...)
In this post
- What is R
- Where did he come from
- Why I love him
- Myths and truth
What's happened R
First of all R is a system for statistical and other scientific calculations using a programming language S
.S- language written by statisticians for statisticians. as defined by author John Chambers. The language has been very well received since its introduction and has been tested by generations of very fastidious statistical users. It can be considered that it is quite widely known and accepted in the global statistical community. On the tongue S A number of critical epidemiological, environmental and financial models have been implemented and are still in operation around the world and in many industries. How is the language from the point of view of me, as a “writing user”, S represents a very pleasant alternative to the language SAS .
From my own experience - Introduction and first lessons S I myself received in the early 90s from WHO statistical experts with whom I crossed paths scientific research of that time.
According to many estimates, R (both for me and not much exaggerated) is one of the most successful open source projects, distributed freely from dozens of mirrors around the world according to GNU license standards.
The authors categorically refuse all proposals to commercialize the project, although today there is reason to assume that the number of installed copies of R in the world exceeds the total number of copies everyone other statistical analysis systems.
From the very beginning to this day, the project evokes in me the deepest respect (on the verge of admiration) for stability, user support, code compatibility, etc., which I would combine in the concept culture.
However, the last sentence is rather for subsequent subsections.
Where did it come from? S and what does this have to do with R
Undoubtedly, Wikipedia will give you many more letters. I will only note what I consider important for understanding the place of S and R in this life in this world.
Bell Laboratories (aka Bell Labs, AT&T Bell Labaratories) are quite famous in the history of science and technology, and IT in particular. Statistical research there was always carried out very seriously and was also seriously supported by all available computer tools (read - tons of Fortran and Lisp code).
What later became the S language originated in the 1970s, at the initiative and direction of John Chambers, as a set of scripts that made it easier to “feed” data to Fortran code. Those. The main focus was on the task of interactive data manipulation, compactness, ease of writing and readability of the code and obtaining decent output on a variety of devices, tables and graphs.
The syntax of the language provides for the construction of almost arbitrarily complex data structures, tools for describing specific statistical tasks and objects - statistics. tests, models, etc.
Since 1984, the language has acquired a name, its own “Bible” (a book by Chambers and Beckers was published: S: An Interactive Environment for Data Analysis and Graphics), began to contain by default an almost complete “gentleman’s set” of statistics and “probability” - distributions, random number generators, statistical tests, many standard statistical analyzes, working with matrices, etc., not to mention the developed system of scientific graphics. The most important thing is that it has become available to users around the world at a very reasonable price.
In 1988 (another book published The New S Language) - modified using OOP, everything became objects with very reasonable default values, accessibility for modification, self-documentation elements, etc., etc.
At the same time, the laboratories published the source code and “Bell Lab” S became free for students and for scientific use. This was all somehow connected with the “dekulakization” of AT&T, but I was no longer very interested in these details.
There were, and probably still are, commercial implementations of the language S. I came across S-Plus And S2000. They were supported by different companies at different times, mainly living (living?) due to the support of previously created S applications. In these post-Bell versions S appeared new version OOP engine, but for a pure user it was almost bloodless in terms of compatibility of historical code.
R- the only non-commercial, completely independent (from the original Bell) implementation of the language S.
And in a rare agreement these days, in some way unimaginable to me, the developers of the current versions of the commercial S and non-profit R support their almost complete compatibility and continuity.
And now R
Behind any significant phenomenon in this life there is some kind of charismatic personality. However, this can happen and this is the definition of the significance of the phenomenon.
In the case of R, there are three such people.
I already mentioned John Chambers.
Ross Ihaka, a student and then a researcher in the Department of Statistics at the University of Auckland, chose the topic of his dissertation (which was carried out at MIT, USA) to study the possibility of building a virtual machine (VM) for statistical programming languages. The intermediate language chosen was Lisp (Common Lisp, CL) and it implements a prototype VM that “understands” small subsets of SAS And S.
Ross returned to Oakland to complete his dissertation, where he soon met Robert Gentleman and became interested in the R project.
Ross never defended his dissertation, but already has academic degree from several universities “based on combined merit.” Last year he was awarded the title and received the position of Associate Professor (Assistant Professor) at his home university.
Robert Gentleman, another statistician with a passion for programming, originally from Canada, while on an internship at the University of Auckland (he was then working in Australia), suggested that Ross “write some language.”
According to the legend that I myself heard from these “founding fathers”, in just almost a month they, in a fit of insane enthusiasm, rewrote C.L. almost all teams S, including a powerful linear modeling library.
Computing engine R, following the traditions of the prototype, the well-known, generally accepted and free BLAS library was chosen (with the ability to use ATLAS, etc. with the same interface).
Paul Murrel, one of Ross's closest friends and also an employee of the University of Oakland, went out of his way and wrote (in C, it seems) a graphics engine from scratch that completely reproduces the functionality of that in S.
The result was a free, fully functional package that instantly gained a place in educational process Oakland University, fully consistent with the descriptions in Chambers’s very detailed and high-quality books, which were traditionally published in paperbacks and of average print quality, but were cheap and accessible.
Several activist groups in the GNU (eg GIS) movement have adopted R as a platform for scientific computing.
But truly the widest fame R acquired in bioinformatics when one of the “fathers” Robert Gentleman, who was involved at that time in the work of the company Affimmetrix, duplicated all the functionality of the company’s commercial software and launched (well, more than one, of course) the open source project Bioconductor. Currently Bioconductor is the undisputed leader in bioinformatics open source for all “omics” (genomics, proteomics, metabolomics, etc.).
Naturally, the single interface language for this riot of bioinformatics fantasies has become R.
The circle came full circle when the retired Chambers, the creator of the language S, became a full member of the group of active developers R.
Why I love him (list)
- Interactivity, “Programming with data” - my favorite style of work
- Elegant (for an amateur) language - I love lists, data frames, functional programming and lambda functions (a-la) Freedom of expression: the same problem can be solved in ten ways (mitigates the feeling of routine)
- “Looks at this world soberly” - rarely “crashes” or “suspends” anyone, logical operations with missing data, error handling at runtime (try-error), easy exchange with the system at the level of standard I/O, etc.
- A complete set of ready-to-use statistical procedures
- Well documented and well maintained - compatibility, continuity, etc.
- Gathered around me a humanly pleasant professional community (forums, user conferences, etc.)
- Well-documented interface for external libraries and functions on anything - Fortran, C, Java. Hence a sea of well-documented libraries on all aspects of statistics and data processing in almost every field of science, but with a primary focus on bioinformatics/biostatistics; everything is updated regularly and correctly, if there is the author’s will for it
- Lack of a mandatory GUI in the “basic configuration” - Well, I’m not a “mouse” person!
That's what I'm actually trying to show in my article.
Why and how I use it (examples)
I started writing in this section, but stopped. Otherwise I would never have finished.
Oh, probably sometime later.
Myths and truth
R slow
R is “thin”, uses blas/lapack/atlas libraries for calculations, try to write something faster than these good old Fortran (often) “workhorses”. All critical functions, as a rule, use vector operations and are implemented in WITH.R uses computing resources irrationally, in particular memory
Yes, the developers admit this sin. But working hours a specialist is now more expensive than hardware. Unload toys from a modern work computer and you will have no problems with R with most real data sets.Free software may not be reliable
Maybe: Fortran, Linux, C, Lisp, Java etc.Instead of an Epilogue
As stated above, the post below is actually a translation of my presentation for a fairly specific target audience, and I will briefly describe this audience.Many “clean” IT companies will have to meet such people, since food production has long been competing with oil and other energy resources to attract capital and generate profits. And the capacity of the bioinformatics market in medicine and pharmacology is limited, no matter how you look at it.
So, my audience is people with a basic education in genetics and breeding, veterinary medicine, and, less often, biology (mainly molecular). Guys and aunties (more of the latter), 20-30 years old... programming (!) on FORTRANe or VB, famously managing Excel tables with 100k rows/columns and periodically “dropping” their Linux computing 500+core 12TB cluster with their tasks (and their programming) shared memory and from time to time requiring expansion of disk memory by another ten terabytes.
The methodological basis is an explosive mixture of variance analyzes as ancient as the world with mixed models solved in no other way than by the maximum likelihood method, “brain-melting” Bayesian networks, etc.
Data - tables of data from units to tens of thousands of rows, sometimes including 1-5 columns with phenotypes, but more and more often - tens or hundreds of “Ka” columns of variables that are weakly correlated with each other and with phenotypes.
Well, yes, they also have a “good tradition” of looking at everything in terms of family ties (genetics, after all). Kinship ties are traditionally presented in the form of a matrix of “kinship ties” (pedigree) with dimensions of, for example, 40,000 x 40,000 (this is if there are 40,000 animals). Well, or (for now, fortunately, only in the project) 20,000,000 x 20,000,000 - this is to “cover” with a single model all 20 million historical animals available in the database ( DB2, if anyone is interested, and even Cobol has not yet been “cut out” from everywhere...)
On tables littered with literature on (at the same time) Fortran, Java, C#, Scala, Octavia, Linux for Dummies you can recognize recent bioinformatics graduates. But somehow quickly many of them leave science to become “coders.”
However, I also know a case of reverse movement. So R will be useful to many more.
Recently I came across such a phenomenon - many people have heard about the R programming language. But very few people know what it is.
Since I am a native speaker of this language and am interested in its popularization, I will try to cover the topic a little in this post. It will be interesting!
The plan is simple:
1) What is the R language
2) Popularity in Russia
What is the R languageR (wiki) is a programming language for statistical data processing and graphics, and a free, open-source computing environment under the GNU Project.
In our opinion: The language is ideal for searching for market patterns. Free, fast and free.
It allows you to conduct statistical studies of everything you can get your hands on. Over the years of its existence, dozens and hundreds of extensions have appeared to solve almost any applied problems.
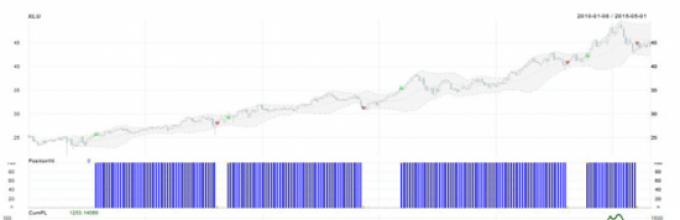
This is THE MOST popular language programming algorithmic traders in the West. This is at the cutting edge of what is currently available in machine learning and statistics. 
In order to write programs in R, just download R-Studio and that’s it

Some time ago, as part of popularizing the language, I recorded a mini-course “R for everyone.” Free. Look how easy it all works and how quickly you can write scripts on it. This is simply great!
Popularity in RussiaLeaves much to be desired.
If in the West 90% of all research is carried out using this language, then in Russia a rare week passes on SmartLab, in which you can find at least one mention of it. Those. the number of people who know R is critically small.
Among the reasons for this situation: the lack of manuals in Russian until recently, the lack of training courses, the low literacy of traders and much more. Let's not talk about this.
The only important thing is that the popularity of this language in Russia and the CIS is steadily growing.
Russian users are tired of broken APIs and expensive trading systems. Free language R, in the context of trading, will very soon take its rightful place in their toolkit. Join us!
That's all for today. I hope it was interesting.
Watch my free course to see how easy it is to write code. Write robots!
Many examples and articles on this site actively use the “R” programming language.
R for Linux can usually be installed using your distribution's repositories. I use Debian-based distributions, where the standard command to install R looks like this:
Sudo apt install r-base
You need to type this in the terminal.
The beauty of R is this:
- This program is free (distributed under the GPL license),
- Many packages have been written for this program to solve wide spectrum tasks. All of them are also free.
- The program is very flexible: the sizes of any vectors and matrices can be changed at the user's request; the data does not have a rigid structure. This property turns out to be extremely useful in the case of forecasting, when the researcher needs to give a forecast for an arbitrary period.
The latter property is especially relevant since other statistical packages (such as SPSS, Eviews, Stata) assume that we may only be interested in analyzing data that has a fixed structure (for example, all data in a working file must be of the same periodicity with the same start dates and end).
However, R is not the friendliest program. While working with it, forget about the mouse - almost all the most important actions in it are performed using the command line. However, in order to make life a little easier, and the program itself a little more welcoming, there is a frontend program called RStudio. You can download it from here. It is installed after R itself has already been installed. RStudio has many convenient tools and a nice interface, however, analysis and forecasting in it are still carried out using the command line.
Let's try to take a look at this wonderful program.
Getting to know RStudio
The RStudio interface looks like this:
In the upper right corner in RStudio the name of the project is indicated (which for now we have “None” - that is, it is missing). If you click on this inscription and select “New Project” ( new project), then we will be asked to create a project. For basic forecasting purposes, just select “New Directory” (a new folder for the project), “Empty Project” (an empty project), and then enter the name of the project and select the directory in which to save it. Use your imagination and try to come up with a name yourself :).
When working with one project, you can always access the data, commands and scripts stored in it.
On the left side of the RStudio window is the console. This is where we will enter various commands. For example, let's write the following:
This command will generate 100 random variables from a normal distribution with zero mathematical expectation and unit variance, after which it will create a vector called “x” and write the resulting 100 values into it. Symbol "<-» эквивалентен символу «=» и показывает какое значение присвоить нашей переменной, стоящей слева. Иногда вместо него удобней использовать символ «->”, although our variable in this case should be on the right. For example, the following code will create an object "y" that is absolutely identical to the object "x":
These vectors now appear in the upper right part of the screen, under the tab, which I have entitled “Environment”:

Changes in the "Environment" tab
This part of the screen will display all the objects that we save during the session. For example, if we create a matrix like this:
\(A = \begin(pmatrix) 1 & 1 \\ 0 & 1 \end(pmatrix) \)
with this command:
then it will appear in the “Environment” tab:

Any function we use requires us to assign some values to certain parameters. The matrix() function has the following parameters:
- data – vector with data that should be written to the matrix,
- nrow – number of rows in the matrix,
- ncol – number of columns in the matrix,
- byrow - logical parameter. If “TRUE” (true), then the matrix will be filled in rows (from left to right, row by row). By default, this parameter is set to FALSE.
- dimnames - a sheet with row and column names.
Some of these parameters have default values (for example, byrow=FALSE ), while others can be omitted (for example, dimnames ).
One of the tricks of “R” is that any function (for example, our matrix() ) can be accessed by specifying values directly:
Or you can do it the way we did it earlier - observing the sequence and omitting the names of the parameters.
In order to see the contents of any object located in the “Environment” tab, just print its name in the console:
Another option is to click on the object name in the "Environment" tab.
where matrix is the name of the function we are interested in. In this case, RStudio will open the “Help” panel especially for you with a description:

You can also find help on a function by typing the name of the function in the “search” window (icon with a lens) in the “Help” tab.
If you don’t remember exactly how to write the name of a function or what parameters are used in it, just start writing its name in the console and press the “Tab” button:

In addition to all this, you can write scripts in RStudio. You may need them if you need to write a program or call a sequence of functions. Scripts are created using the button with a plus sign in the upper left corner (you need to select “R Script” in the drop-down menu). In the window that opens after this, you can write any functions and comments. For example, if we want to build line graph along row x, this can be done as follows:
Plot(x) lines(x)
The first function builds a simple scatter plot, and the second function adds lines on top of the points connecting the points sequentially. If you select these two commands and press “Ctrl+Enter”, they will be executed, causing RStudio to open the “Plot” tab in the lower right corner and display the plotted plot in it.

If we still need all the typed commands in the future, then this script can be saved (floppy disk in the upper left corner).
In case you need to refer to a command that you have already typed sometime in the past, there is a “History” tab at the top right of the screen. In it you can find and select any command you are interested in and double-click to paste it into the console. In the console itself, you can access previous commands using the Up and Down buttons on your keyboard. The “Ctrl+Up” key combination allows you to show a list of all recent commands in the console.
In general, RStudio has a lot of useful keyboard shortcuts that make working with the program much easier. You can read more about them.
As I mentioned earlier, there are many packages for R. All of them are located on the CRAN server and to install any of them you need to know its name. Installation and updating of packages is carried out using the “Packages” tab. By going to it and clicking on the “Install” button, we will see something like the following menu:

Let's type in the window that opens: forecast is a package written by Rob J. Hyndman, containing a bunch of functions useful to us. Click the “Install” button, after which the “forecast” package will be installed.
Alternatively, we can install any package, knowing its name, using the command in the console:
Install.packages("smooth")
provided that it is, of course, in the CRAN repository. smooth is a package whose functions I develop and maintain.
Some packages are only available in source codes on sites like github.com and require that they be compiled first. To build packages under Windows, you may need the Rtools program.
To use any of the installed packages, you need to enable it. To do this, you need to find it in the list and tick it, or use the command in the console:
Library(forecast)
One unpleasant problem may appear in Windows: some packages are easily downloaded and assembled, but are not installed in any way. R in this case writes something like: "Warning: unable to move temporary installation...". All you need to do in this case is to add the folder with R to the exceptions in your antivirus (or turn it off while installing packages).
After downloading the package, all the functions included in it will be available to us. For example, the tsdisplay() function, which can be used like this:
Tsdisplay(x)
She will build us three graphs, which we will discuss in the chapter “Forecaster Toolkit”.
In addition to the forecast package, I often use the Mcomp package for various examples. It contains data series from the M-Competition database. Therefore, I recommend that you install it too.
Very often we will need not just data sets, but data of the “ts” class (time series). In order to make a time series from any variable, you need to run the following command:
Here the start parameter allows us to specify the date from which our time series begins, and frequency specifies the frequency of the data. The number 12 in our example indicates that we are dealing with monthly data. As a result of executing this command, we transform our vector “x” into a time series of monthly data starting from January 1984.
These are the basic elements of R and RStudio that we will need.
P.P.S. Enough good guide on R published on the QSAR4U website by Pavel Polishchuk.
P.P.P.S. A lot of information about R is presented on the official website.
Independent work
Here are some tasks for you to do yourself in R. Run the following commands, see what happens and try to understand why this happened:
(41/3 + 78/4)*2 2^3+4 1/0 0/0 max(1,min(-2.5),max(2,pi)) sqrt(3^2+4^2 ) exp(2)+3i log(1024) log(1024, base=2) c(1:3) c(1:5)*2 + 4 x
In August 1993, two young New Zealand scientists from the University of Auckland announced their new development, which they called R. According to the creators, Robert Gentleman and Ross Ihaka, it was supposed to be a new implementation of the S language, differing from S-PLUS in some details, for example, the handling of global and local variables, as well as working with memory. In fact, they did not create a complete analogue of S-PLUS, but a new “branch” on the “S tree”. Many of the things that distinguish R from S-PLUS are due to the influence of the Scheme language (a functional programming language, one of the most popular dialects of the Lisp language).
By mid-2016, R had caught up with SAS and SPSS (which are paid) and became one of the three most common systems for processing statistical information. It should also be noted that R is one of the 10 general purpose programming languages.
Possibilities
Many statistical methods are implemented in the R environment: linear and nonlinear models, statistical hypothesis testing, time series analysis, classification, clustering, graphical visualization. The R language allows you to define your own functions. Many R functions are written in R itself. For computational complex tasks It is possible to implement functions in C, C++ and Fortran. Advanced users can directly access R objects from C code. R is a more rigorous object-oriented language than most statistical computing languages. Graphics functions allow you to create graphs of good printing quality, with the ability to include mathematical symbols. It has its own LaTeX-like documentation format.
Although R is most often used for statistical computing, it can also be used as a matrix computing tool. Like MATLAB, R treats the result of any number operation as a vector of unit length. Generally speaking, there are no scalars in R.
Scripts
Simply opening an R session and entering commands into the program window, one after another, is just one of the possible ways to work. A much more productive method, which is also a major advantage of R, is the creation of scripts (programs), which are then loaded into R and interpreted by it. From the very beginning of work, you should create scripts, even for tasks that seem trivial - this will significantly save time in the future. Creating scripts for any reason and even without a special reason is one of the foundations of the work culture in R.
Packages
Another important advantage of R is the availability of numerous extensions or packages for it. Some basic packages are present immediately after installing R on the computer, without them the system simply does not work (for example, a package called base, or the grDevices package, which controls the output of graphs), as well as “recommended” packages (a package for specialized cluster analysis cluster, a package for the analysis of nonlinear models nlme and others). In addition, you can install any of the almost eight thousand (as of mid-2016) packages available on CRAN. If you have Internet access, this can be done directly from R using the install.packages() command.
Links
- CRAN (Comprehensive R Archive Network) - central system storage and distribution of R and its packages.
]. "Proletarian of mental labor." Physicist by education. I have been working in the field of processing medical and biological information for 30+ years.
I’ve been working in R for exactly 10 years, having migrated to it after 15 years of close collaboration with Matlab. The primary reason for migrating to another work platform was my own physical migration to the opposite end of the Earth in Auckland, New Zealand. Here life from the first days pushed me into the arms of R, which I have not yet had to regret.
More and more often I see outbreaks of interest in R in the professional network. Well, articles about him appear on this respected resource. Further below the cut is my first attempt at a Russian-language introduction to R - the first (verbal) part of the presentation that I did for colleagues from the Animal Science Faculty, Iova State University three years ago.
(to the side: but it turns out that it’s difficult to translate yourself...)
In this post
- What is R
- Where did he come from
- Why I love him
- Myths and truth
What's happened R
First of all R is a system for statistical and other scientific calculations using a programming language S
.S- language written by statisticians for statisticians. as defined by author John Chambers. The language has been very well received since its introduction and has been tested by generations of very fastidious statistical users. It can be considered that it is quite widely known and accepted in the global statistical community. On the tongue S A number of critical epidemiological, environmental and financial models have been implemented and are still in operation around the world and in many industries. How is the language from the point of view of me, as a “writing user”, S represents a very pleasant alternative to the language SAS .
From my own experience - Introduction and first lessons S I myself received it in the early 90s from WHO statistical experts with whom I intersected on scientific research at that time.
According to many estimates, R (both for me and not much exaggerated) is one of the most successful open source projects, distributed freely from dozens of mirrors around the world according to GNU license standards.
The authors categorically refuse all proposals to commercialize the project, although today there is reason to assume that the number of installed copies of R in the world exceeds the total number of copies everyone other statistical analysis systems.
From the very beginning to this day, the project evokes in me the deepest respect (on the verge of admiration) for stability, user support, code compatibility, etc., which I would combine in the concept culture.
However, the last sentence is rather for subsequent subsections.
Where did it come from? S and what does this have to do with R
Undoubtedly, Wikipedia will give you many more letters. I will only note what I consider important for understanding the place of S and R in this life in this world.
Bell Laboratories (aka Bell Labs, AT&T Bell Labaratories) are quite famous in the history of science and technology, and IT in particular. Statistical research there was always carried out very seriously and was also seriously supported by all available computer tools (read - tons of Fortran and Lisp code).
What later became the S language originated in the 1970s, at the initiative and direction of John Chambers, as a set of scripts that made it easier to “feed” data to Fortran code. Those. The main focus was on the task of interactive data manipulation, compactness, ease of writing and readability of the code and obtaining decent output on a variety of devices, tables and graphs.
The syntax of the language provides for the construction of almost arbitrarily complex data structures, tools for describing specific statistical tasks and objects - statistics. tests, models, etc.
Since 1984, the language has acquired a name, its own “Bible” (a book by Chambers and Beckers was published: S: An Interactive Environment for Data Analysis and Graphics), began to contain by default an almost complete “gentleman's set” of statistics and “probabilistic scientists” - distributions, random number generators, statistical tests, many standard statistical analyzes, working with matrices, etc., not to mention a developed system of scientific graphics. The most important thing is that it has become available to users around the world at a very reasonable price.
In 1988 (another book published The New S Language) - modified using OOP, everything became objects with very reasonable default values, accessibility for modification, self-documentation elements, etc., etc.
At the same time, the laboratories published the source code and “Bell Lab” S became free for students and for scientific use. This was all somehow connected with the “dekulakization” of AT&T, but I was no longer very interested in these details.
There were, and probably still are, commercial implementations of the language S. I came across S-Plus And S2000. They were supported by different companies at different times, mainly living (living?) due to the support of previously created S applications. In these post-Bell versions S a new version of the OOP engine appeared, but for the pure user it was almost bloodless in terms of compatibility of the historical code.
R- the only non-commercial, completely independent (from the original Bell) implementation of the language S.
And in a rare agreement these days, in some way unimaginable to me, the developers of the current versions of the commercial S and non-profit R support their almost complete compatibility and continuity.
And now R
Behind any significant phenomenon in this life there is some kind of charismatic personality. However, this can happen and this is the definition of the significance of the phenomenon.
In the case of R, there are three such people.
I already mentioned John Chambers.
Ross Ihaka, a student and then a researcher in the Department of Statistics at the University of Auckland, chose the topic of his dissertation (which was carried out at MIT, USA) to study the possibility of building a virtual machine (VM) for statistical programming languages. The intermediate language chosen was Lisp (Common Lisp, CL) and it implements a prototype VM that “understands” small subsets of SAS And S.
Ross returned to Oakland to complete his dissertation, where he soon met Robert Gentleman and became interested in the R project.
Ross never defended his dissertation, but already has an academic degree from several universities “on the basis of combined merit.” Last year he was awarded the title and received the position of Associate Professor (Assistant Professor) at his home university.
Robert Gentleman, another statistician with a passion for programming, originally from Canada, while on an internship at the University of Auckland (he was then working in Australia), suggested that Ross “write some language.”
According to the legend that I myself heard from these “founding fathers”, in just almost a month they, in a fit of insane enthusiasm, rewrote C.L. almost all teams S, including a powerful linear modeling library.
Computing engine R, following the traditions of the prototype, the well-known, generally accepted and free BLAS library was chosen (with the ability to use ATLAS, etc. with the same interface).
Paul Murrel, one of Ross's closest friends and also an employee of the University of Oakland, went out of his way and wrote (in C, it seems) a graphics engine from scratch that completely reproduces the functionality of that in S.
The result was a free, fully functional package that instantly gained a place in the educational process at Oakland University, fully consistent with the descriptions in Chambers's very detailed and high-quality books, which were traditionally published in paperbacks and of average print quality, but were cheap and accessible.
Several activist groups in the GNU (eg GIS) movement have adopted R as a platform for scientific computing.
But truly the widest fame R acquired in bioinformatics when one of the “fathers” Robert Gentleman, who was involved at that time in the work of the company Affimmetrix, duplicated all the functionality of the company’s commercial software and launched (well, more than one, of course) the open source project Bioconductor. Currently Bioconductor is the undisputed leader in bioinformatics open source for all “omics” (genomics, proteomics, metabolomics, etc.).
Naturally, the single interface language for this riot of bioinformatics fantasies has become R.
The circle came full circle when the retired Chambers, the creator of the language S, became a full member of the group of active developers R.
Why I love him (list)
- Interactivity, “Programming with data” - my favorite style of work
- Elegant (for an amateur) language - I love lists, data frames, functional programming and lambda functions (a-la) Freedom of expression: the same problem can be solved in ten ways (mitigates the feeling of routine)
- “Looks at this world soberly” - rarely “crashes” or “suspends” anyone, logical operations with missing data, error handling at runtime (try-error), easy exchange with the system at the level of standard I/O, etc.
- A complete set of ready-to-use statistical procedures
- Well documented and well maintained - compatibility, continuity, etc.
- Gathered around me a humanly pleasant professional community (forums, user conferences, etc.)
- Well-documented interface for external libraries and functions on anything - Fortran, C, Java. Hence a sea of well-documented libraries on all aspects of statistics and data processing in almost every field of science, but with a primary focus on bioinformatics/biostatistics; everything is updated regularly and correctly, if there is the author’s will for it
- Lack of a mandatory GUI in the “basic configuration” - Well, I’m not a “mouse” person!
That's what I'm actually trying to show in my article.
Why and how I use it (examples)
I started writing in this section, but stopped. Otherwise I would never have finished.
Oh, probably sometime later.
Myths and truth
R slow
R is “thin”, uses blas/lapack/atlas libraries for calculations, try to write something faster than these good old Fortran (often) “workhorses”. All critical functions, as a rule, use vector operations and are implemented in WITH.R uses computing resources irrationally, in particular memory
Yes, the developers admit this sin. But a specialist’s working time is now much more expensive than hardware. Unload toys from a modern work computer and with most real data sets you will have no problems with R.Free software may not be reliable
Maybe: Fortran, Linux, C, Lisp, Java etc.Instead of an Epilogue
As stated above, the post below is actually a translation of my presentation for a fairly specific target audience, and I will briefly describe this audience.Many “clean” IT companies will have to meet such people, since food production has long been competing with oil and other energy resources to attract capital and generate profits. And the capacity of the bioinformatics market in medicine and pharmacology is limited, no matter how you look at it.
So, my audience is people with a basic education in genetics and breeding, veterinary medicine, and, less often, biology (mainly molecular). Guys and aunties (more of the latter), 20-30 years old... programming (!) on FORTRANe or VB, famously managing Excel tables with 100k rows/columns and periodically “dropping” their tasks (and their programming) on their Linux computing 500+ core cluster with 12TB of shared memory and from time to time requiring the expansion of disk memory by another ten terabytes.
The methodological basis is an explosive mixture of variance analyzes as ancient as the world with mixed models solved in no other way than by the maximum likelihood method, “brain-melting” Bayesian networks, etc.
Data - tables of data from units to tens of thousands of rows, sometimes including 1-5 columns with phenotypes, but more and more often - tens or hundreds of “Ka” columns of variables that are weakly correlated with each other and with phenotypes.
Well, yes, they also have a “good tradition” of looking at everything in terms of family ties (genetics, after all). Kinship ties are traditionally presented in the form of a matrix of “kinship ties” (pedigree) with dimensions of, for example, 40,000 x 40,000 (this is if there are 40,000 animals). Well, or (for now, fortunately, only in the project) 20,000,000 x 20,000,000 - this is to “cover” with a single model all 20 million historical animals available in the database ( DB2, if anyone is interested, and even Cobol has not yet been “cut out” from everywhere...)
On tables littered with literature on (at the same time) Fortran, Java, C#, Scala, Octavia, Linux for Dummies you can recognize recent bioinformatics graduates. But somehow quickly many of them leave science to become “coders.”
However, I also know a case of reverse movement. So R will be useful to many more.